Decoding the RNA m6A modification in human early embryonic development

Photo: Guro Flor Lien, OUH
A Powerful Tool: picoMeRIP-seq
RNA modifications are tiny chemical marks added to RNA molecules to control how they work. One of the most common is m6A modification, which acts like a “sticky note” on RNA—telling the cell when to use it, store it, or discard it. These marks help fine-tune RNA activity during important processes like fertilization and the very early stages of embryo.
In this study, the Klungland group used their latest cutting-edge, ultra-sensitive technique called picoMeRIP-seq [1], which can detect m6A even from just 10 cells. This allowed them to draw the first complete map of m6A across human eggs and early embryos—from immature eggs (GV) to mature eggs (MII), zygotes, and all the way to blastocysts. They found around 5,000 m6A-marked genes at each stage, mostly marked near the stop codon—the place where the RNA stops when coding for proteins. These tags followed a typical sequence pattern known as RRACH.
Humans and Mice: Same Blueprint, Different Patterns
To understand how universal these RNA tags are, the team compared their human data to their earlier published mouse data [2]. Surprisingly, they found substantial differences. During a critical stage called zygotic genome activation (ZGA)—when the embryo switches from using mom’s RNAs to using its own—mice had far more m6A-marked genes than humans. It’s like two artists working from the same sketch but choosing different color palettes. This suggests that the differences in RNA marking may have key roles on how embryos develop to different species.
Maternal Messages and Ancient Echoes
Before fertilization, a mother loads helpful RNA “instructions” into the egg—like packing a suitcase for a long journey. These maternal RNAs are essential for starting development but need to be removed at the right time. Many RNAs were marked with m6A, possibly as a signal for when to be removed. They also found m6A on transposons—old genetic elements from ancient viruses, like HERVH, that are active in early development. When the team blocked METTL3, the enzyme that adds m6A to RNA, the levels of these transposon RNAs dropped. This suggests that m6A may help regulate these unusual RNAs that quietly shape early life.
Why It Matters: A Window into Life’s First Days
According to the WHO, infertility impacts approximately 1 in 6 couples globally. Assisted reproductive technologies, like IVF, offer hope—but many embryos don’t develop into healthy babies. To improve this, we need to understand how early embryos work at the molecular level. This is the first study to chart RNA m6A modification across all early stages of human embryo development. It lays the foundation for exploring how RNA chemical signals guide the very early stages of existence—and may guide to improve success rates in fertility treatments in the future.
This study was a joint effort between the University of Oslo, Oslo University Hospital, and the University of Michigan.
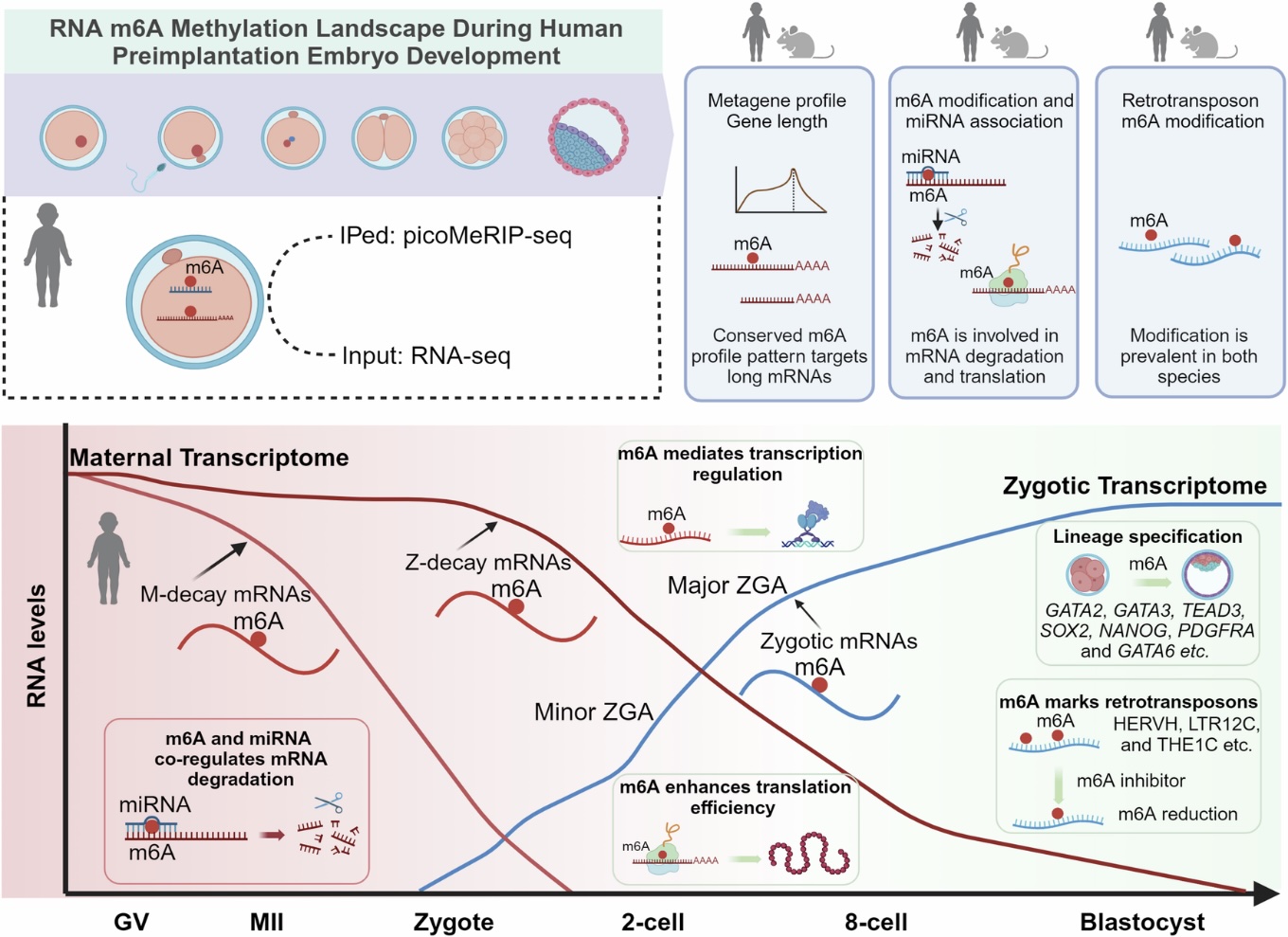
Figure is from the paper.
Read the full paper, published in The EMBO Journal, June 4, 2025::
The RNA m6A landscape during human oocyte-to-embryo transition | The EMBO Journal
Reference:
1. Li, Y., et al., Single-cell m(6)A mapping in vivo using picoMeRIP-seq. Nat Biotechnol, 2024. 42(4): p. 591-596.
2. Wang, Y., et al., The RNA m(6)A landscape of mouse oocytes and preimplantation embryos. Nat Struct Mol Biol, 2023. 30(5): p. 703-709.
Links:
Yanjiao Li - publications
Laboratory for Dynamic Gene Regulation research group, led by Arne Klungland
Department of Microbiology, OUH
Centre for Embryology and Healthy Development (CRESCO) - Health in the earliest stage of life
News article from the CRESCO home page:
Decoding the RNA m6A modification in human early embryonic development (cresco.uio.no)
