Per Olaf Ekstrøm
- Senior scientist; Dr. philos.
- +47 2278 1847
Background
|
I was introduced to research in 1991 when I start working at The Norwegian Radium hospital (DNR) as a research technician. Parallel to this work I was able to complete my Ph.D. with the title “The use of microdialysis to investigate extracellular pharmacokinetics of methotrexate” in 1997. This was followed by a 2-year post doc period at MIT. Here I work with separating DNA by capillary electrophoresis and thouched upon US cancer mortality curves as shown on Prof. Thillys web page. In 1999 I returned to DNR and continued the development of DNA separation in multi capillary instruments. This lead to the first publication on the method of separation of DNA by cycling temperature capillary electrophoresis (December 2002, (PMID: 14599347)). Optimized and explored the separation principles of cycling temperature capillary electrophoresis which resulted in more than a dozen publications (PMID: 18600220, 22539319). Given the intimate knowledge of the method and the instrumentation, I can be regarded as an expert in this field. In the course of my research career I have embraced methods, methods development and technical skill on chemistry, troubleshooting and instrument maintains. Four Ph.D students have graduated under my supervision and used cycling temperature capillary electrophoresis (CTCE) as their main analysis method. |
|
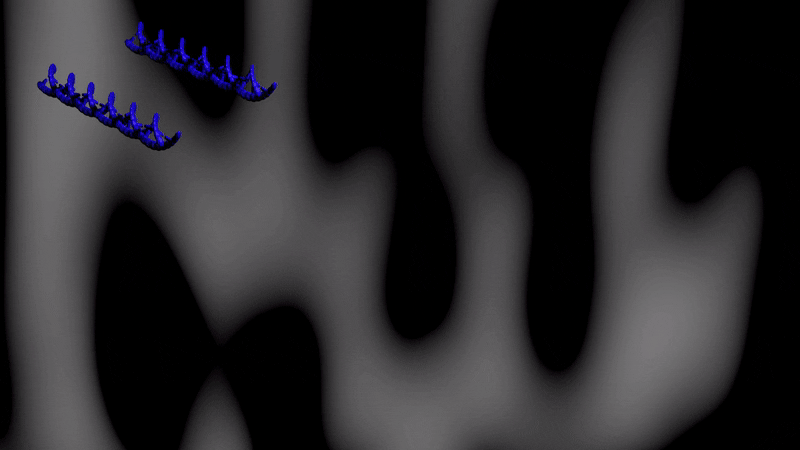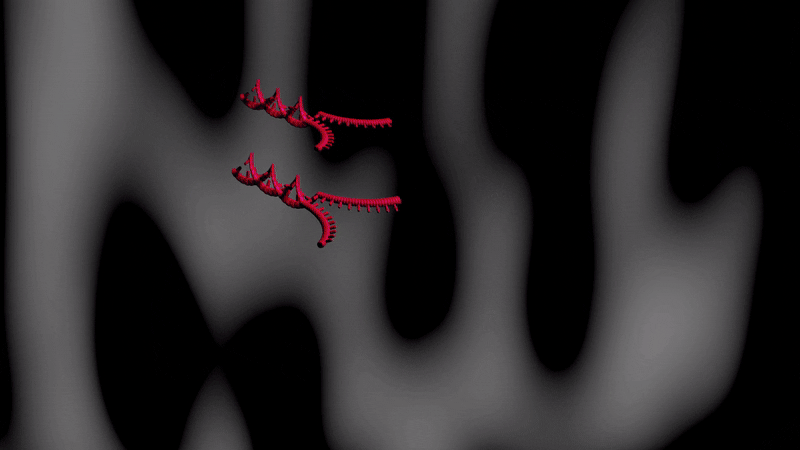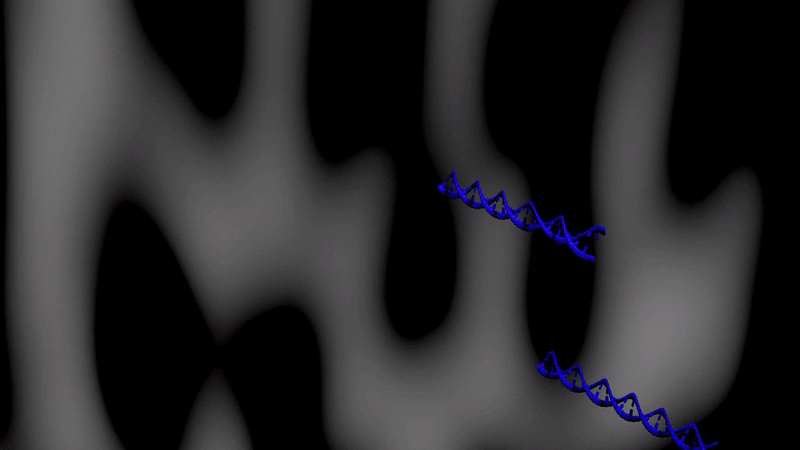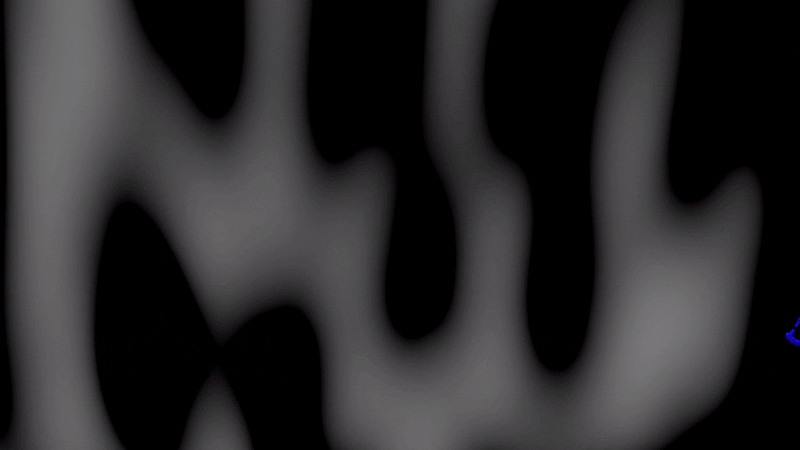
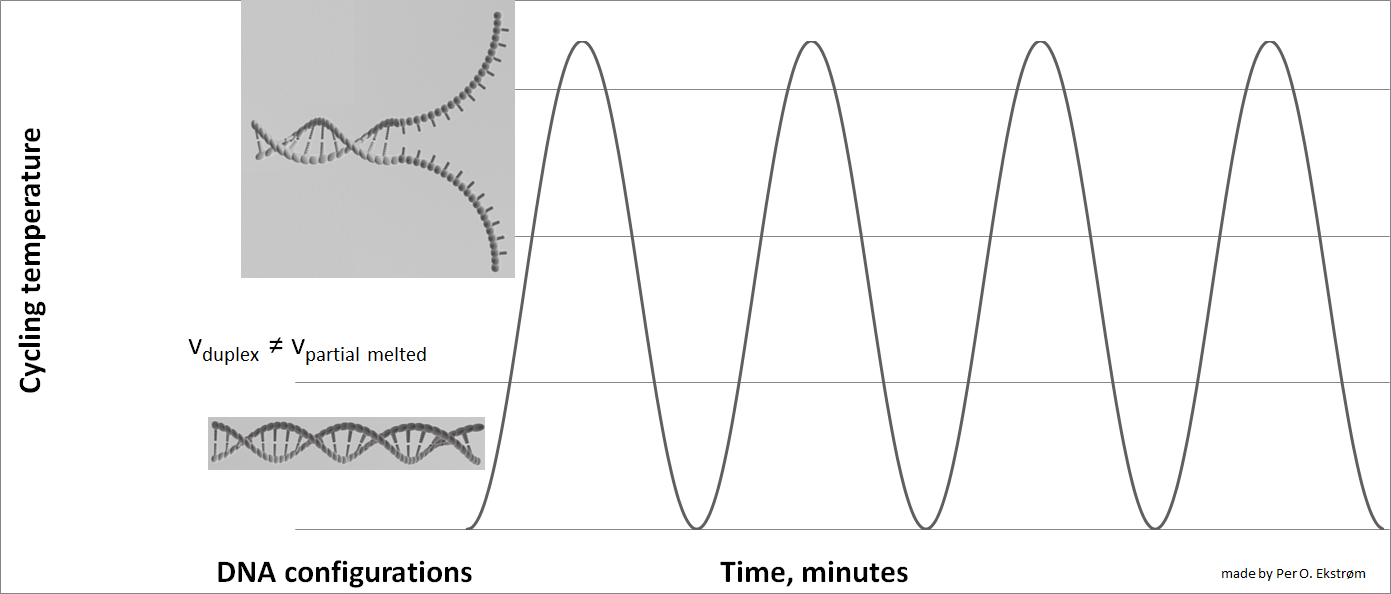
Below is an illustartion of separation by CTCE of two DNA molecules with the same length and one base pair difference. The separation takes place during one temperature cycle, in a capillary filled with an appropriate sieving matrix while the DNA migrates in an electric field (electrophoresis). The more cycles applied the better separation between the DNA variants.
|
Effect of cycling temperature on DNA Color change indicate temperature change (blue-cold, red=warmer)
|
Velocity (v) of DNA is dependent on the entanglemet with the sieving matrix during electrophoresis.
|
Projects
| Distribution of mitochondrial mutant fractions within tumors. 3D-tumor |
Instrumentation, Cycling temperature capillary electrophoresis (CTCE). MegaBACE 1000 resource page |
|
Distribution of somatic mutant fractions within a mouse tumor model. |
Testing of in-house chemistry for Sars-Cov-2 analysis. |
|
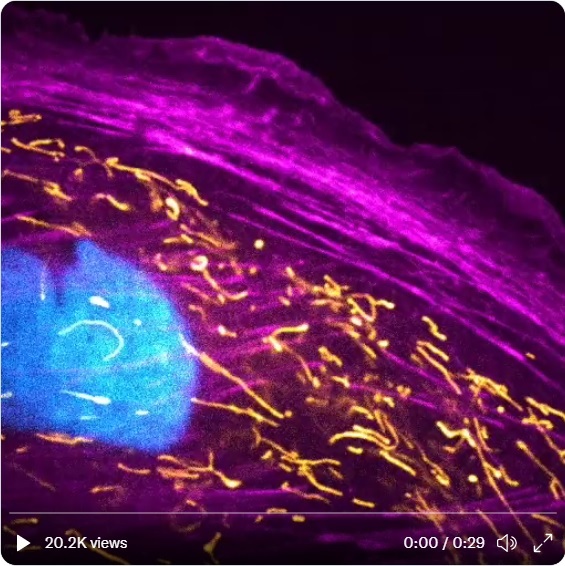
For information Mitocohndrial research, the struckture usally depicted as a bean but it is NOT.
A cell was videoed through a confocal scanning microscope. DNA in the nucleus (blue), the powerhouses/overlords of the cell, mitochondria (yellow), and actin filaments (purple) are shown. |
Quantification of mitochondrial copy numbers with in-house chemistry (Work in progress)
Us of mitochondrial DNA variants as marker for clonality (Work in progress)
|
Publications 2023
Rapid assessment of 3-dimensional intra-tumor heterogeneity through cycling temperature capillary electrophoresis
BMC Res Notes, 16 (1), 167
DOI 10.1186/s13104-023-06437-5, PubMed 37568187
Publications 2022
Genetic Characterization in High-Risk Individuals from a Low-Resource City of Peru
Cancers (Basel), 14 (22)
DOI 10.3390/cancers14225603, PubMed 36428697
Publications 2021
The Quandary of DNA-Based Treatment Assessment in De Novo Metastatic Prostate Cancer in the Era of Precision Oncology
J Pers Med, 11 (5)
DOI 10.3390/jpm11050330, PubMed 33922147
Publications 2020
Somatic Mitochondrial DNA Point Mutations Used as Biomarkers to Demonstrate Genomic Heterogeneity in Primary Prostate Cancer
Prostate Cancer, 2020, 7673684
DOI 10.1155/2020/7673684, PubMed 32908706
Tracing of Human Tumor Cell Lineages by Mitochondrial Mutations
Front Oncol, 10, 523860
DOI 10.3389/fonc.2020.523860, PubMed 33344219
Publications 2019
Results of multigene panel testing in familial cancer cases without genetic cause demonstrated by single gene testing
Sci Rep, 9 (1), 18555
DOI 10.1038/s41598-019-54517-z, PubMed 31811167
Publications 2018
Genetic variants of prospectively demonstrated phenocopies in BRCA1/2 kindreds
Hered Cancer Clin Pract, 16, 4
DOI 10.1186/s13053-018-0086-0, PubMed 29371908
Potentially pathogenic germline CHEK2 c.319+2T>A among multiple early-onset cancer families
Fam Cancer, 17 (1), 141-153
DOI 10.1007/s10689-017-0011-0, PubMed 28608266
Identification of genetic variants for clinical management of familial colorectal tumors
BMC Med Genet, 19 (1), 26
DOI 10.1186/s12881-018-0533-9, PubMed 29458332
Publications 2017
Is detection of intraperitoneal exfoliated tumor cells after surgical resection of rectal cancer a prognostic factor of survival?
BMC Cancer, 17 (1), 406
DOI 10.1186/s12885-017-3365-7, PubMed 28592327
Mapping mitochondrial heteroplasmy in a Leydig tumor by laser capture micro-dissection and cycling temperature capillary electrophoresis
BMC Clin Pathol, 17, 6
DOI 10.1186/s12907-017-0042-3, PubMed 28405177
Quantifying mitochondrial DNA copy number using robust regression to interpret real time PCR results
BMC Res Notes, 10 (1), 593
DOI 10.1186/s13104-017-2913-1, PubMed 29132417
Publications 2016
Scanning the mitochondrial genome for mutations by cycling temperature capillary electrophoresis
Mitochondrial DNA A DNA Mapp Seq Anal, 29 (1), 19-30
DOI 10.1080/24701394.2016.1233532, PubMed 27728990
Tp53/p53 status in keratoacanthomas
J Cutan Pathol, 43 (7), 571-8
DOI 10.1111/cup.12713, PubMed 27020606
Cycling temperature capillary electrophoresis: A quantitative, fast and inexpensive method to detect mutations in mixed populations of human mitochondrial DNA
Mitochondrion, 29, 65-74
DOI 10.1016/j.mito.2016.04.006, PubMed 27166160
Publications 2015
Automated amplicon design suitable for analysis of DNA variants by melting techniques
BMC Res Notes, 8, 667
DOI 10.1186/s13104-015-1624-8, PubMed 26559640
Ten modifiers of BRCA1 penetrance validated in a Norwegian series
Hered Cancer Clin Pract, 13 (1), 14
DOI 10.1186/s13053-015-0035-0, PubMed 26052370
Publications 2014
TP53/p53 alterations and Aurora A expression in progressor and non-progressor colectomies from patients with longstanding ulcerative colitis
Int J Mol Med, 35 (1), 24-30
DOI 10.3892/ijmm.2014.1974, PubMed 25333414
Publications 2013
Influence of polymorphisms in genes encoding immunoregulatory proteins and metabolizing enzymes on susceptibility and outcome in patients with diffuse large B-cell lymphoma treated with rituximab
Leuk Lymphoma, 54 (10), 2205-14
DOI 10.3109/10428194.2013.774392, PubMed 23391141
Publications 2012
Separation principles of cycling temperature capillary electrophoresis
Electrophoresis, 33 (7), 1162-8
DOI 10.1002/elps.201100550, PubMed 22539319
Polymorphisms in genes encoding interleukin-10 and drug metabolizing enzymes GSTP1, GSTT1, GSTA1 and UGT1A1 influence risk and outcome in Hodgkin lymphoma
Leuk Lymphoma, 53 (10), 1934-44
DOI 10.3109/10428194.2012.682307, PubMed 22475179
Publications 2010
Validation and use of microdialysis for determination of pharmacokinetic properties of the chemotherapeutic agent mitomycin C - an experimental study
BMC Cancer, 10, 469
DOI 10.1186/1471-2407-10-469, PubMed 20809961
Publications 2008
Scanning for DNA variants by denaturant capillary electrophoresis
Methods Mol Biol, 439, 19-34
DOI 10.1007/978-1-59745-188-8_2, PubMed 18370093
Analysis of mutational spectra by denaturing capillary electrophoresis
Nat Protoc, 3 (7), 1153-66
DOI 10.1038/nprot.2008.79, PubMed 18600220
Molecular detection (k-ras) of exfoliated tumour cells in the pelvis is a prognostic factor after resection of rectal cancer?
BMC Cancer, 8, 213
DOI 10.1186/1471-2407-8-213, PubMed 18655729
Fetal-juvenile origins of point mutations in the adult human tracheal-bronchial epithelium: absence of detectable effects of age, gender or smoking status
Mutat Res, 646 (1-2), 25-40
DOI 10.1016/j.mrfmmm.2008.08.016, PubMed 18824180
Publications 2007
Technology to accelerate pangenomic scanning for unknown point mutations in exonic sequences: cycling temperature capillary electrophoresis (CTCE)
BMC Genet, 8, 54
DOI 10.1186/1471-2156-8-54, PubMed 17697348
Frequency of TP53 mutations in relation to Arg72Pro genotypes in non small cell lung cancer
Cancer Epidemiol Biomarkers Prev, 16 (10), 2077-81
DOI 10.1158/1055-9965.EPI-07-0153, PubMed 17932356
Publications 2006
Evaluation of sieving matrices used to separate alleles by cycling temperature capillary electrophoresis
Electrophoresis, 27 (10), 1878-85
DOI 10.1002/elps.200500642, PubMed 16619298
Coding region polymorphisms in T cell signal transduction genes. Prevalence and association to development of multiple sclerosis
J Neuroimmunol, 177 (1-2), 40-5
DOI 10.1016/j.jneuroim.2006.04.021, PubMed 16764945
Association of a functional polymorphism in the promoter of the MDM2 gene with risk of nonsmall cell lung cancer
Int J Cancer, 119 (3), 718-21
DOI 10.1002/ijc.21872, PubMed 16496380
Publications 2005
Review of denaturant capillary electrophoresis in DNA variation analysis
Electrophoresis, 26 (13), 2520-30
DOI 10.1002/elps.200410403, PubMed 15934053
BRAF mutation detection and identification by cycling temperature capillary electrophoresis
Electrophoresis, 26 (13), 2553-61
DOI 10.1002/elps.200410427, PubMed 15948220
High-throughput gender determination using automated denaturant gel capillary electrophoresis
Electrophoresis, 26 (13), 2562-6
DOI 10.1002/elps.200410392, PubMed 15934052
Detection and frequency estimation of rare variants in pools of genomic DNA from large populations using mutational spectrometry
Mutat Res, 570 (2), 267-80
DOI 10.1016/j.mrfmmm.2004.11.004, PubMed 15708585
Lack of association with the CD28/CTLA4/ICOS gene region among Norwegian multiple sclerosis patients
J Neuroimmunol, 166 (1-2), 197-201
DOI 10.1016/j.jneuroim.2005.06.002, PubMed 16005527
Publications 2004
Determination of hereditary mutations in the BRCA1 gene using archived serum samples and capillary electrophoresis
Anal Chem, 76 (15), 4406-9
DOI 10.1021/ac049788k, PubMed 15283579
Publications 2003
Approach to analysis of single nucleotide polymorphisms by automated constant denaturant capillary electrophoresis
Mutat Res, 526 (1-2), 75-83
DOI 10.1016/s0027-5107(03)00033-2, PubMed 12714185
Direct identification of all oncogenic mutants in KRAS exon 1 by cycling temperature capillary electrophoresis
Electrophoresis, 24 (1-2), 63-9
DOI 10.1002/elps.200390032, PubMed 12652573
DNA variants in the ATM gene are not associated with sporadic rectal cancer in a Norwegian population-based study
Int J Colorectal Dis, 19 (1), 49-54
DOI 10.1007/s00384-003-0519-7, PubMed 12827413
Cycling gradient capillary electrophoresis: a low-cost tool for high-throughput analysis of genetic variations
Electrophoresis, 24 (11), 1716-22
DOI 10.1002/elps.200305384, PubMed 12783447
Publications 2002
Mutation detection in KRAS Exon 1 by constant denaturant capillary electrophoresis in 96 parallel capillaries
Anal Biochem, 304 (2), 200-5
DOI 10.1006/abio.2002.5629, PubMed 12009696
Reducing the cost of consumables used for ABI 310 genetic analyzer
Anal Biochem, 306 (1), 148-9
DOI 10.1006/abio.2002.5656, PubMed 12069426
Population screening of single-nucleotide polymorphisms exemplified by analysis of 8000 alleles
J Biomol Screen, 7 (6), 501-6
DOI 10.1177/1087057102238623, PubMed 14599347
Detection of mutations in exon 8 of TP53 by temperature gradient 96-capillary array electrophoresis
Biotechniques, 33 (3), 650-3
DOI 10.2144/02333pf01, PubMed 12238774
Publications 2001
Automated constant denaturant capillary electrophoresis applied for detection of KRAS exon 1 mutations
Biotechniques, 30 (5), 972-5
DOI 10.2144/01305st01, PubMed 11355359
Mutation analysis of TP53 exons 5-8 by automated constant denaturant capillary electrophoresis
Tumour Biol, 22 (5), 323-7
DOI 10.1159/000050634, PubMed 11553863
Publications 2000
Two-point fluorescence detection and automated fraction collection applied to constant denaturant capillary electrophoresis
Biotechniques, 29 (3), 582-4, 586-9
DOI 10.2144/00293rr01, PubMed 10997272
Publications 1999
Detection of low-frequency mutations in exon 8 of the TP53 gene by constant denaturant capillary electrophoresis (CDCE)
Biotechniques, 27 (1), 128-34
DOI 10.2144/99271rr01, PubMed 10407675
Publications 1997
The use of microdialysis to investigate extracellular pharmacokinetics of methotrexate
University of Oslo, Oslo, 1 b. (flere pag.)
BIBSYS 972142363, ISBN 82-7722-076-6
Continuous intratumoral microdialysis during high-dose methotrexate therapy in a patient with malignant fibrous histiocytoma of the femur: a case report
Cancer Chemother Pharmacol, 39 (3), 267-72
DOI 10.1007/s002800050571, PubMed 8996531
Intratumoral differences in methotrexate levels within human osteosarcoma xenografts studied by microdialysis
Life Sci, 61 (19), PL275-80
DOI 10.1016/s0024-3205(97)00839-4, PubMed 9364204
Alterations in methotrexate pharmacokinetics by naproxen in the rat as measured by microdialysis
Life Sci, 60 (24), PL 359-64
DOI 10.1016/s0024-3205(97)00241-5, PubMed 9188769
Publications 1996
Determination of extracellular methotrexate tissue levels by microdialysis in a rat model
Cancer Chemother Pharmacol, 37 (5), 394-400
DOI 10.1007/s002800050403, PubMed 8599860
Publications 1995
Mikrodialyse av metotrexat i en rottemodell: en teknikk for bestemmelse av et cytostatikums vevsdistribusjon
[P.O. Ekstrøm>, Oslo, 1 b. (flere pag.)
BIBSYS 951032674
Pharmacokinetics of different doses of methotrexate at steady state by in situ microdialysis in a rat model
Cancer Chemother Pharmacol, 36 (4), 283-9
DOI 10.1007/BF00689044, PubMed 7628046