Selected research output
 |
Addressing the challenge of transcriptomic heterogeneity of primary colorectal cancers: We show that intra-tumor heterogeneity of the consensus molecular subtypes is frequent and associated with a poor patient survival independently of the tumor microenvironment (Nat Commun 2024). A classification with less vulnerability to tumor heterogeneity and with improved prognostic power was identified from genes with uniform expression patterns among multiregional tumor samples. However, evidence of phenotypic plasticity of cancer cells between matched primary tumors and metastatic lesions emphasized the challenge of reconciliation of primary and metastatic tumor classifications. |
 |
The need for metastasis-oriented transcriptomic classifications of colorectal cancer: We implemented approaches to adjust for gene expression signals from the tumor microenvironment in cancer tissue samples, and developed a computational tool for unbiased classification of preclinical models (Sci Rep 2017) and liver metastases (NPJ Genom Med 2021) according to the consensus molecular subtypes. The tool is available as the R package CMScaller. Based on detection of pronounced metastatic heterogeneity of the subtypes, we performed de novo subtype discovery of metastatic lesions (Genome Med 2021). This showed the potential for a clinically relevant transcriptomic classification in the context of metastatic tumor heterogeneity. |
 |
Modeling subclonal evolution of cancer metastasis: We reconstruct the clonal architectures and subclonal evolution of primary and metastatic tumors based on their mutational profiles. In a study of ovarian cancer (JCI Insight 2024), we showed frequent polyclonal dissemination within the peritoneal cavity. Dissemination occurred as a single and rapid wave of clones with relatively low mutational diversity, except in cancers exposed to chemotherapy. Cancers with DNA damage repair deficiency disseminated particularly late in their evolutionary trajectory models. In ongoing studies we map the subclonal evolutionary patterns of metastasis of colorectal cancers. |
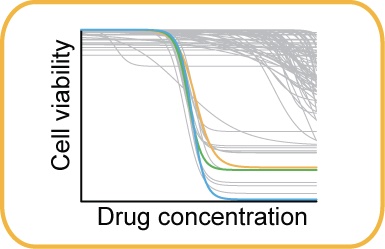 |
Treatment modeling: We conduct pharmacogenomic studies by integrating drug screens of tumor organoids and cancer cell lines with molecular profiles. We showed potential to overcome resistance to chemotherapy in a particularly aggressive subtype of colorectal cancer using a targeted combination therapy (Clin Cancer Res 2018). We identified TP53 wild-type activity as a mechanism of response to PARP inhibition in colorectal cancer (EBioMed 2020). We also demonstrated that the allele-specific expression level of mutations has impact on their interpretation as cancer biomarkers (Genome Med 2021). |
