Department of Genetics scientists with publication in PNAS
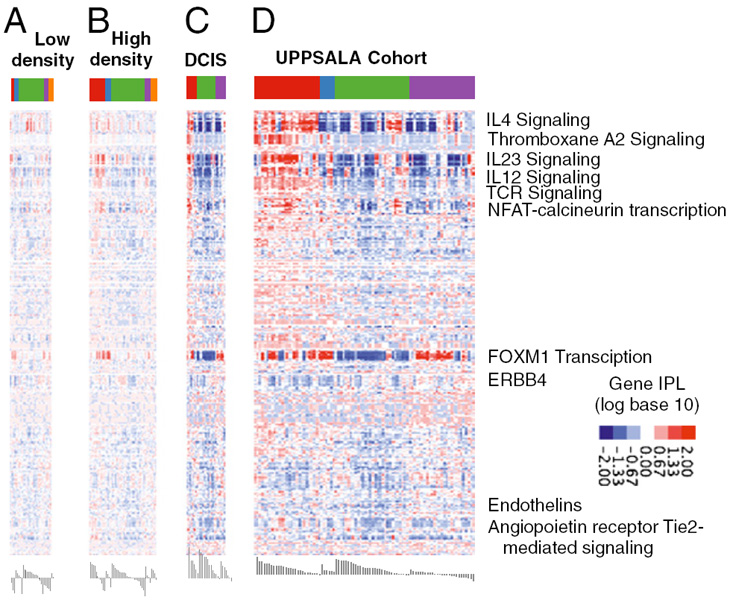
In this paper they show that combining mRNA expression and DNA copy number can classify the patients in groups that provide the best predictive value with respect to prognosis and identified key molecular and stromal signatures. One such is the chronic inflammatory signature, which promotes the development and/or progression of various epithelial tumors.
They could demonstrate that within the adaptive immune lineage, the strongest predictor of good outcome is the acquisition of a gene signature that favors a high T-helper 1 (Th1)/cytotoxic T-lymphocyte response at the expense of Th2-driven humoral immunity.
Patients who have breast cancer with a basal HER2-negative molecular profile (PDGM2) are characterized by high expression of protumorigenic Th2/humoral-related genes (2438%) and a low Th1/Th2 ratio. The luminal molecular subtypes are again differentiated by low or high FOXM1 and ERBB4 signaling. these distinct interleukin signaling profiles observed in
invasive cancers were absent or weakly expressed in healthy tissue but already prominent in ductal carcinoma in situ (DCIS), together with ECM and cell-cell adhesion regulating pathways. Differences were observed also in healthy breast tissue between low and high mammographic density, the most prominent difference being in STAT4 signaling.
Links
Integrated molecular profiles of invasive breast tumors and ductal carcinoma in situ (DCIS) reveal differential vascular and interleukin signaling. (link to PubMed)
Kristensen VN, Vaske CJ, Ursini-Siegel J, Van Loo P, Nordgard SH, Sachidanandam R, Sørlie T, Wärnberg F, Haakensen VD, Helland A, Naume B, Perou CM, Haussler D, Troyanskaya OG, Børresen-Dale AL.
Proc Natl Acad Sci U S A. 2011 Sep 9. [Epub ahead of print]
Home page of Vessela N. Kristensen's group - Cancer genome variation
Home page of Anne-Lise Børresen-Dale's group - Molecular Biology of Breast Cancer
Børresen-Dale's CV and publications
