Qualitative transcript variation in cancer
A lot of information on the behaviour of cancers can be read from the RNA. In addition to quantitative changes in RNA levels, gene expression is influenced by qualitative changes. Much of these changes are due to alternative and aberrant splicing, promoter usage, and fusion genes. In fact, several types of cancers vary in their genome-wide extent of aberrant pre-mRNA splicing. The phenomenon, named transcriptome instability (TIN), was reported as a novel phenotype in colorectal cancer as well as in prostate and other types of cancer (Sveen et al., 2011 and 2014). Interestingly, TIN is linked to the expression level of a large set of splicing factors.
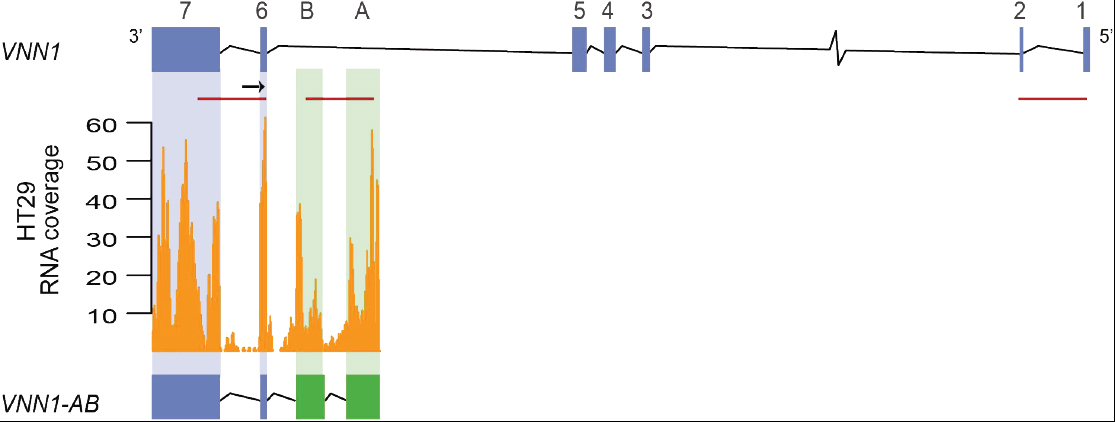
The research group have discovered several transcript structures relevant to cancer. These include splice variants in colorectal cancer (Figure; Løvf et al., 2014) and fusion transcripts in colorectal (Nome et al., 2013), prostate (Zhao et al., 2017), and testicular cancers (Hoff et al., 2016). All these novel transcript variants have all been discovered with the use of innovative bioinformatics strategies from paired-end RNA-sequencing and whole-genome sequencing data.
Selected publications
Strømme JM, Johannessen B, Kidd SG, Bogaard M, Carm KT, Zhang X, Sveen A, Mathelier A, Lothe RA, Axcrona U, Axcrona K, and Skotheim RI (2022). Expressed prognostic biomarkers for primary prostate cancer independent of multifocality and transcriptome heterogeneity. Cancer Gene Ther. 29(8-9): 1276-84
Carm KT, Hoff AM, Bakken AC, Axcrona U, Axcrona K, Lothe RA, Skotheim RI*, and Løvf M (2019). The clinical usefulness of molecular classification of primary prostate cancer is challenged by interfocal heterogeneity. Sci. Rep. 9: 13579 *Corresponding author
Hoff AM, Alagaratnam S, Zhao S, Bruun J, Andrews PW, Lothe RA, and Skotheim RI (2016). Identification of novel fusion genes in testicular germ cell tumors. Cancer Res. 76(1): 108-16
Hoff AM*, Johannessen B*, Alagaratnam S, Zhao S, Nome T, Løvf M, Bakken AC, Hektoen M, Sveen A, Lothe RA, and Skotheim RI (2015). Novel RNA-variants in colorectal cancers. Oncotarget 6(34): 36587-602 *Equal contribution
Løvf M, Nome T, Bruun J, Eknæs M, Bakken AC, Mpindi JP, Kilpinen S, Rognum TO, Nesbakken A, Kallioniemi O, Lothe RA, Skotheim RI (2014). A novel transcript, VNN1-AB, as a biomarker for colorectal cancer. Int. J. Cancer 135(9):2077-84
Nome T, Thomassen GOS, Bruun J, Ahlquist TC, Bakken AC, Rognum T, Nesbakken A, Lorenz S, Sun J, Barros-Silva JD, Lind GE, Myklebost O, Teixeira MR, Meza-Zepeda LA, Lothe RA, and Skotheim RI (2013). Common fusion transcripts identified in colorectal cancer cell lines by high throughput RNA sequencing. Transl. Oncol. 6(5):546-53
Alagaratnam S, Harrison NJ, Bakken AC, Hoff A, Jones M, Sveen A, Moore H, Andrews PW, Lothe RA, and Skotheim RI (2013). Transforming pluripotency: an exon-level study of malignancy-specific transcripts in human embryonal carcinoma and embryonic stem cells. Stem Cells Dev. 22(7): 1136-1146
Sveen A, Bakken AC, Ågesen TH, Lind GE, Nesbakken A, Nordgård O, Brackmann S, Rognum TO, Lothe RA, and Skotheim RI (2012). The exon-level biomarker SLC39A14 has organ confined cancer specificity in colorectal cancer. Int. J. Cancer 131: 1479-1485
Sveen A, Ågesen TH, Nesbakken A, Rognum TO, Lothe RA, and Skotheim RI (2011). Transcriptome instability in colorectal cancer identified by exon microarray analyses: Associations with splicing factor expression levels and patient survival. Genome Medicine 3(5): 32