About the centre
Head of the Center: Professor Anne-Lise Børresen-Dale
Host institution: Institute for Cancer Research (ICR), Oslo University Hospital, Radiumhospitalet (OUHR)
Vision: Towards personalized therapy for breast cancer; integrated molecular and epidemiological studies of breast cancer to reduce risk, improve prognosis, and tailor the treatment.
Senior Partners: A-L. Børresen-Dale, prof. PhD, Head of Dep. of Genetics, OUHR. V. Kristensen, prof. PhD, Head of Genome Cancer Variation group OUHR. Ø. Fodstad, prof. MD, PhD, Head of ICR, OUHR. G.M. Mælandsmo, prof. PhD, Head of Dep. of Tumor Biology, OUHR. B. Naume, MD, PhD, Head of Laboratory for studies of Micrometastasis, OUHR. E. Schlichting, MD, PhD, Head of BESD, OUH-Ull. R. Kåresen, prof. emeritus, MD, PhD BESD, OUH-Ull. E. Wist, prof. MD, PhD, Head of Norwegian Breast Cancer Group (NBCG), OUH-Ull. T. Sauer, Prof. MD, PhD Head of gyn/breast pathology OUH-Ull. O. Engebråten, Ass. prof. MD, PhD, OUS-Ull. I. Gribbestad, Prof. PhD, Head of MR cancer group, Dept. Circl. and Med. Imaging, NTNU.
Research aims
The overall aim of the research center is to open up for stronger collaboration between its clinical and basic scientists, and to motivate exchange of ideas on how to best utilize the huge collection of patient materials and data generated for the benefit of the patients. By such synergism we will be able to explore genes/pathways/networks involved in basic processes like cell cycle, DNA repair, apoptosis, and immune response and their impact on breast cancer development, progression and response to therapy. By performing longitudinal studies of samples at different stages of the disease and characterize such patient materials in full molecular details, we aim to develop more individual treatment protocols.
The specific aims of the center is:
- Develop validated stratification criteria based on phenotypic/genotypic profiling of breast tumors.
- Use validated phenotypic and genotypic stratification criteria for assessing individual response and prognosis in patients with breast cancer
- Identify molecular pathways and biomarkers predicting treatment response and/or resistance using cell lines and orthotopic xenograft models representing the various subgroups of breast cancer
- Translate and validate molecular and imaging biomarkers from preclinical models into clinical trials.
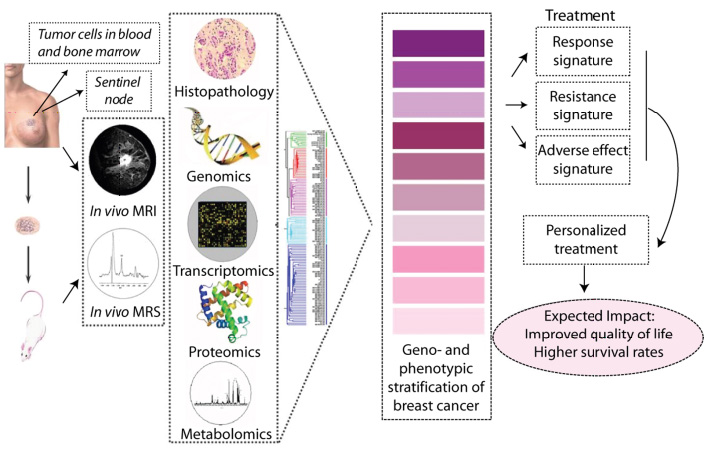
Figure 1. Workflow: “From bed- to bench- to byte- to bed-side”. To ensure an optimal framework for an integrative approach towards individualized therapy for breast cancer matched material from the primary tumor, sentinel lymph nodes, peripheral blood, bone marrow and metastatic lesions are collected. In the presented research program the groups at Oslo University Hospital and that at NTNU, each with high international recognition in their fields, will make joint effort in studies aiming to enhance our knowledge of the molecular characteristics of breast cancer.
Project outline
Activity 1: Global characterization of the primary tumor and patients' genetic background.
Present classification of breast cancer with classical histopathological markers is insufficient as similar tumors often experience different clinical outcome. Tumor aggressiveness and therapeutic response are associated with the combined influence of multiple rather than single genes. Cancer development and progression is a complex biological evolutionary process since tumors are composite organs with dynamical genomes shaped by gene aberrations, epigenetic changes, cellular context, characteristics of the individual patient and environmental influences. High throughput molecular profiling approaches are thus supposed to be more informative, sensitive and specific than single markers to reflect the actual heterogeneity of the disease. We are testing this hypothesis by utilizing several technologies to globally characterize the primary tumor at the genome, transcriptome, proteome and metabolic level, as well as the patients' genetic background, and evaluate the data in an integrated systems biology approach and relate the findings to clinical parameters in the various clinical cohorts.
Biopsies from the primary tumor and the sentinel lymph nodes, as well as blood and bone marrow are collected and presence of micrometastases in blood and bone marrow are monitored at the time of diagnosis. In this activity molecular profiling of primary tumors on multiple levels (mRNA, miRNA, CGH, methylation, protein, SNPs and CNVs) are performed, followed by analyses of the generated data with respect to tumor characteristics, pathological findings, presence of DTC (disseminated tumor cells), disease relapse and response to treatment. The biostatistical data analysis will be performed together with our established collaborators in Norway and abroad. We have accumulated considerable experience with these types of studies from pilot projects and acquired adequate methodological and technical know-how. For several of the collected materials molecular profiling at different levels has been completed, others are ongoing and will continue.
Activity 2: Detection and characterization of occult tumor cells in BM (DTC), blood (CTC) and SLN
To metastasize, cancer cells have to detach from the primary tumor and penetrate into the circulation before they may extravasate and establish growth in a new organ. Little is known about the tumor cell characteristics that determine whether cells in the circulation will die, stay dormant or form a metastasis. Different methods exist for detecting disseminated cancer cells and the methods seem to identify different subpopulations of tumor cells. It is therefore of great interest to characterize the cells and their distinct markers that determine further clinical progression of the disease and also the response to therapy.
Planned activities - anticipated progress and expected achievements: In an ongoing study, occult tumor cells from BM and SLN are detected on the basis of expression of cell surface antigens and further characterized by molecular methods (RT-PCR, aCGH). Results from the molecular analyses demonstrate a remarkable variation in cell-specific expression of genes normally associated with cancer progression (ie.: EMT factors). Results obtained will be compared with those obtained on samples of the primary tumor and the aim is to identify biomarkers that are able to predict the metastatic potential of the patient tumor.
Activity 3. Model systems and functional studies
Breast cancer cell lines and animal models mimicking human cancer provide important fundaments for studies on cancer initiation and progression, as well as for testing potential diagnostics and therapeutic agents. We have access to most commercially available human breast cancer cell lines. Furthermore, material obtained from primary breast cancer biopsies has been implanted in mice and ortothopic growing xenografts, having retained the molecular characteristics of the patient's tumors, have been established. Pre-clinical evaluation in ortothopic- or experimental metastasis models will provide material for molecular investigations of induced effects and the results obtained may assist in the selection of treatments for patients.
Planned activities - anticipated progress and expected achievements: The integrative analyses from "Activity 1 and 2" will suggest signaling pathways, candidate biomarkers and therapeutic targets that will be screened and validated by in vitro models. Multiple targets will be silenced in functional screens; cells will be lysed and printed as Lysate MicroArrays (LMA) and stained with antibodies through our collaborator (Olli Kallioniemi). This enables us to study multiple endpoints from a single functional experiment. Following this initial screen, promising targets will also be over-expressed and the drug responses will be evaluated from a selection of targeted drugs. Cell viability using luminescence and LMAs will be used as read-outs in these screens. By this approach, novel key regulators and drug candidates will be identified and subjected to further testing in the pre-clinical models. Evaluation of well known targeted therapeutics is already ongoing in the pre-clinical models. The harvested material is subjected to molecular analyses as described in "Activity 1" and specific signaling pathways and biomarkers predicting treatment responses are sought identified. Candidate marker molecules will be in silico validated in clinical cohorts. The goal is to identify markers or key regulators/pathways predicting drug response of both well-known cancer drugs as well as novel ones. Further molecular and functional characterization of the identified candidate molecules from the in vitro and in vivo models will depend on the candidate in question. At present the molecular mechanisms and drug response of the following genes are investigated: the EMT-inducer S100A4, the immunomodulatory and drug response protein B7H3, and the epidermal growth factor receptor HER-2. The goal will be to unravel molecular mechanisms, identify key regulators at several levels and drug responses that can be utilized for therapeutic intervention.
Activity 4. Integrated analyses
The accumulation of molecular data at all levels allows for respective molecular classification at each level but also for a combinatorial classification acquiring information from each contributing layer. In other words, to identify cancer subtype specific gene networks that drive the biology of breast cancers, we need to develop methods to simultaneously overlay gene expression and copy number data on protein levels and interactions, with information of transcription factor binding and signaling networks. This will enable us to identify cancer-specific driver networks exploring coincident genomic and transcriptional disturbances in local network neighborhoods. A lot of our present work is dedicated to this effort and many of our recent publications illustrate our progress. However. the search for the most informative parameters in such a composite molecular model has just started and we will continue as a planned activity together with our own group of biostatistics and bioinformatics as well as collaborators in Norway and abroad.
Activity 5. Metabolic and tumor physiological characterization by MR
Metabolic characterization of the primary tumor using high resolution magic angle spinning MR spectroscopy (HR-MAS) will be performed by our collaborators at the MR Metabolomics Lab (NTNU/St. Olavs hospital). In vivo characterization of tumor physiological features like perfusion, flow, extra-cellular extra-vascular space, cell density and blood volume will be available through dynamic contrast enhanced and diffusion MRI. Similar analyses have been performed in vivo using the orthotopic xenografts and ex vivo using biopsy specimens, and this will continue using the more recent collected material (Oslo 2) and additional xenograft models. Recent convincing results demonstrate the usefulness of the orthotopic models. These methods have recently been implemented for high field clinical breast cancer examinations.
