Liquid biopsies in early cancer detection
Aims:
The overall aim has been to develop liquid biopsy-based biomarkers for surveillance of cancer development among high-risk patients. Early detection will qualify patient groups now dying of the disease for curative treatment.
The specific aim has been to identify biomarkers for earlier and more accurate detection of cholangiocarcinoma (CCA) among patients with primary sclerosing cholangitis (PSC). The main goal has been to facilitate timely intervention with improved patient outcomes.
Highlights of a bile-based DNA methylation test for CCA detection
- High accuracy for detecting CCA among high-risk PSC patients
- May detect cancer up to 12 months prior to current detection modalities
- Shows potential for earlier cancer detection which enhances the likelihood of curative treatment
- Are currently used in experimental diagnostics in conjunction with standard clinical practice
Background
CCA is a diverse group of malignancies originating from the biliary duct epithelium, and can be categorized anatomically as intrahepatic (iCCA), perihilar (pCCA) or distal (dCCA) [1]. Although considered a rare malignancy globally, its incidence and mortality has been rising, and only a small percentage of patients survive until 5 years after diagnosis [1]. PSC is a well-established risk factor for CCA, contributing to more than 30% of PSC-related deaths [2]. The lifetime risk of CCA development in PSC is considerable, reaching up to 20%, with an annual incidence of 1.5% (Vedeld et al JHEP Rep 2020) [3].
Early detection of CCA in PSC poses a significant challenge, as the inflammatory biliary strictures characteristic of PSC can mimic early malignant changes. Current diagnostic methods, including imaging, cytological evaluation, and assessment of serum tumor markers, face limitations in accuracy. Consequently, a substantial portion of CCAs in PSC are not diagnosed until liver transplantation or at autopsy [4], negatively impacting on prognosis. Surgery remains the only curative option, primarily for early-stage perihilar tumors. However, most patients are diagnosed at advanced stages which prevent the option of surgical intervention, leading to an overall survival of less than 12 months [4]. Since systemic chemotherapy offers only modest improvements in overall survival, earlier detection through novel biomarkers is crucial for enhancing outcomes.
Identification of DNA methylation markers to accurately detect CCA in PSC
Our extensive and long-standing collaboration with the Norwegian PSC centre has led to the identification of DNA methylation markers with promising diagnostic potential. In 2012, through a genome-wide approach we identified a four-marker panel with high sensitivity (87%) and specificity (100%) for differentiating between CCA and benign liver diseases, including PSC (Andresen el al, Epigenetics 2012) [5] (Figure 1). These and additional markers were further analyzed in biliary brushes from patients with CCA and benign liver diseases, where they obtained similar high accuracy as they did in tissue samples (Andresen et al, Hepatology 2015) [6] (Figure 1).
In a Nordic study from 2022 (Vedeld, Grimsrud et al, Hepatology 2022) [7] we used droplet digital PCR (ddPCR), including an in-house developed pipeline that ensures standardization and reliability of the analyses, to analyze small amounts of bile (200ul) from patients with PSC alone and in combination with CCA. The pipeline included i) the 4Plex (Pharo et al, Clin Epigenetics 2018) [8]; an in- house developed internal reference used for normalizing DNA input, and ii) PodCall (Jeanmougin et al Bioinformatics 2023) [9]; an R package performing automated calling of positive droplets, quantification and normalization of methylation levels. We showed that four DNA methylation markers could identify cancer among PSC patients with high sensitivity (100%) and specificity (90%; Figure 1). Interestingly, the test could identify cancer up to one year before clinically detected, which is highly beneficial as an earlier diagnosis enhances the likelihood of curative treatment (Vedeld, Grimsrud et al, Hepatology 2022) [7].
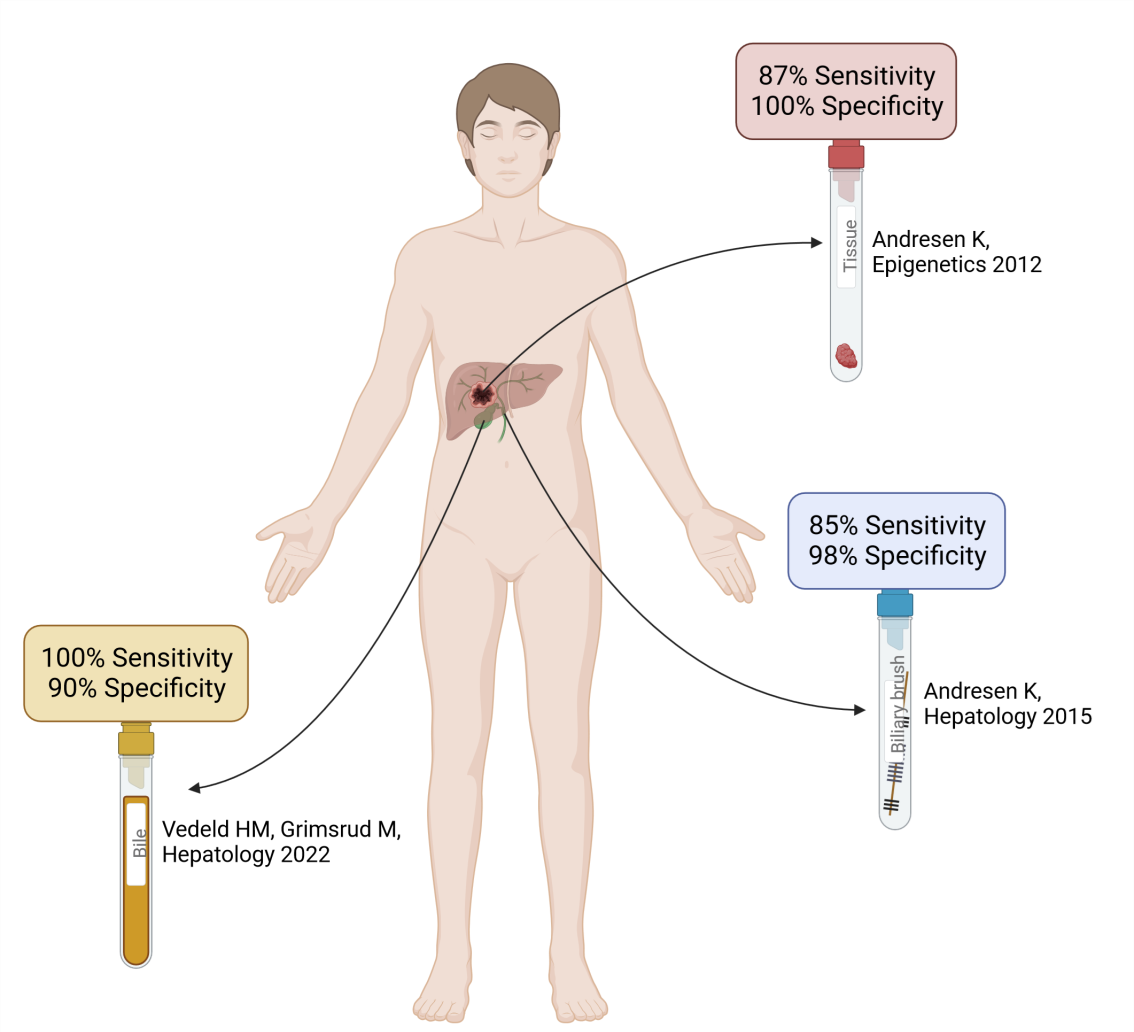
Figure 1. Visualization of the main results from our original research. Created with BioRender.com
Experimental testing of the DNA methylation biomarkers in the clinic
Due to the promising results of the bile test, showing high accuracy for earlier detection of CCA among high- risk PSC patients, the test is currently being used in experimental diagnostics (in conjunction with standard clinical practice).
Pasientsammendrag (på norsk)
Vi forsker på å finne bedre måter for å oppdage kreft i gallegangene (kolangiokarsinom) tidligere hos høyrisiko pasienter (pasienter med primær skleroserende kolangitt (PSC)). Kolangiokarsinom er en sjelden kreftform, men dødeligheten er høy, og risikoen for a utvikle kolangiokarsinom er høyere for de med PSC. Sannsynligheten for å overleve er større dersom kreften oppdages tidlig, ettersom det gir mulighet for kurativ behandling. Det er utfordrende å oppdage kreften, spesielt i pasienter med underliggende PSC, men våre biomarkører viser lovende resultater. Med disse biomarkørene, som kan finnes igjen i små mengder med galle, kan vi oppdage kolangiokarsinom i pasienter med PSC et helt år før dagens metoder. Testen brukes nå til eksperimentell diagnostikk sammen med standard klinisk praksis. Håpet er at testen skal kunne stille en tidligere kreftdiagnose blant pasienter med PSC, og dermed forbedre behandlingsmulighetene og sannsynligheten for å overleve. Vår forskningsgruppe samarbeider med det norske PSC-senteret ved Rikshospitalet.
References
[1] J. M. Banales, J. J. G. Marin, A. Lamarca, P. M. Rodrigues, S. A. Khan, L. R. Roberts, et al., "Cholangiocarcinoma 2020: the next horizon in mechanisms and management," Nature Reviews Gastroenterology & Hepatology, vol. 17, pp. 557-588, 2020/09/01 2020.
[2] K. Boonstra, R. K. Weersma, K. J. van Erpecum, E. A. Rauws, B. W. Spanier, A. C. Poen, et al., "Population-based epidemiology, malignancy risk, and outcome of primary sclerosing cholangitis," Hepatology, vol. 58, pp. 2045-55, Dec 2013.
[3] H. M. Vedeld, T. Folseraas, and G. E. Lind, "Detecting cholangiocarcinoma in patients with primary sclerosing cholangitis - The promise of DNA methylation and molecular biomarkers," JHEP Rep, vol. 2, p. 100143, Oct 2020.
[4] K. M. Boberg, A. Bergquist, S. Mitchell, A. Pares, F. Rosina, U. Broomé, et al., "Cholangiocarcinoma in primary sclerosing cholangitis: risk factors and clinical presentation," Scand J Gastroenterol, vol. 37, pp. 1205-11, Oct 2002.
[5] K. Andresen, K. M. Boberg, H. M. Vedeld, H. Honne, M. Hektoen, C. A. Wadsworth, et al., "Novel target genes and a valid biomarker panel identified for cholangiocarcinoma," Epigenetics, vol. 7, pp. 1249-57, Nov 2012.
[6] K. Andresen, K. M. Boberg, H. M. Vedeld, H. Honne, P. Jebsen, M. Hektoen, et al., "Four DNA methylation biomarkers in biliary brush samples accurately identify the presence of cholangiocarcinoma," Hepatology, vol. 61, pp. 1651-9, May 2015.
[7] H. M. Vedeld, M. M. Grimsrud, K. Andresen, H. D. Pharo, E. von Seth, T. H. Karlsen, et al., "Early and accurate detection of cholangiocarcinoma in patients with primary sclerosing cholangitis by methylation markers in bile," Hepatology, vol. 75, pp. 59-73, Jan 2022.
[8] H. D. Pharo, K. Andresen, K. C. G. Berg, R. A. Lothe, M. Jeanmougin, and G. E. Lind, "A robust internal control for high-precision DNA methylation analyses by droplet digital PCR," Clin Epigenetics, vol. 10, p. 24, 2018.
[9] M. Jeanmougin, H. P. Brodal, H. Dietrichson Pharo, H. M. Vedeld, and G. E. Lind, "PoDCall: positive droplet calling and normalization of droplet digital PCR DNA methylation data," Bioinformatics, vol. 39, Jan 1 2023.
