
Christin Lund-Andersen
- Researcher; PhD
- +47 22 78 17 61
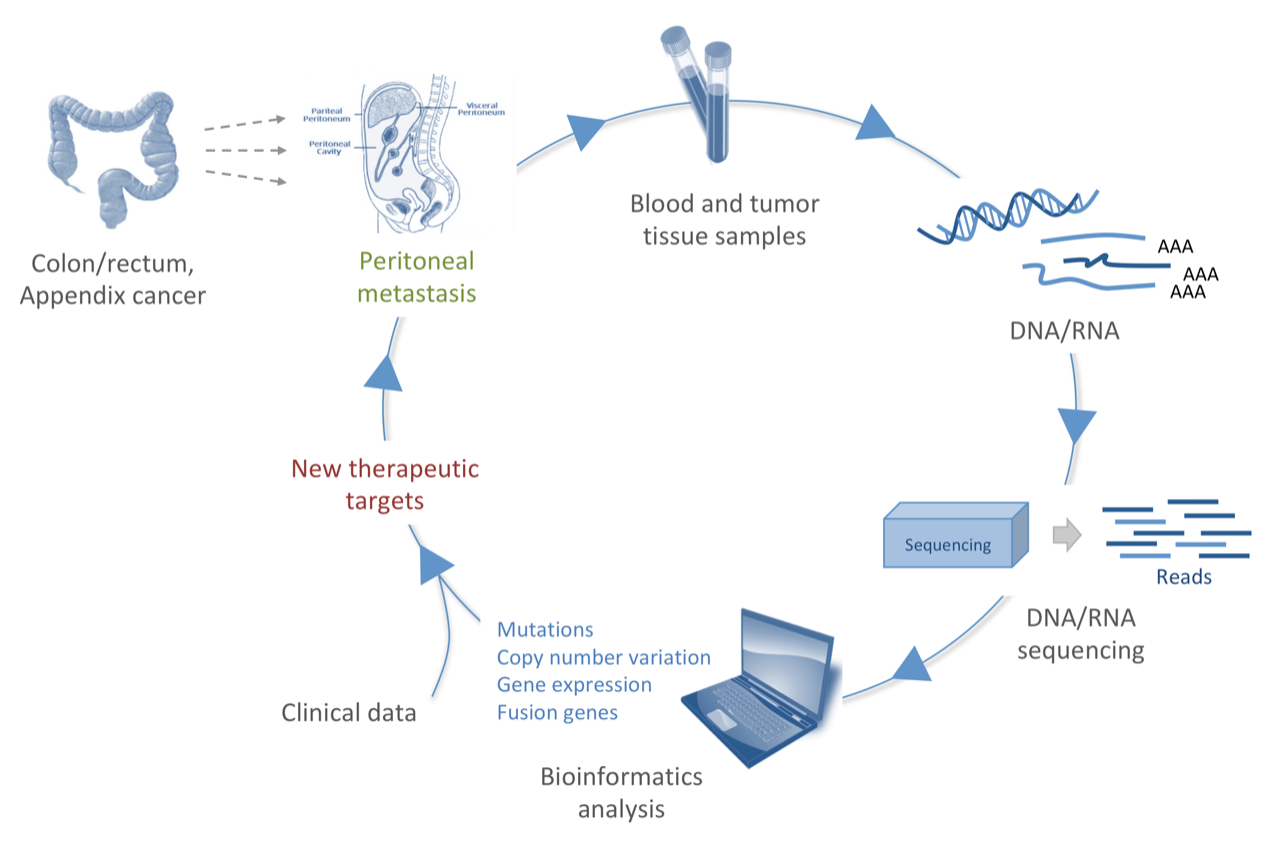
Peritoneal metastasis (PM) is characterized by the presence of widespread tumor lesions on the peritoneal surfaces, and is a major cause of cancer death in abdominal malignancies. The peritoneum is the second most common site for metastatic colorectal cancer (CRC) after liver, and is implicated in at least 25-30% of recurrences. Patients with PM-CRC have a poor prognosis (median survival of 30 months), and are more resistant to chemotherapy compared to other metastatic sites such as liver and lung. PM originating from appendiceal tumors (pseudomyxoma peritonei, PMP), on the other hand, has a better prognosis, where current treatment may cure up to 50% of the patients. PMP is a rare disease characterized by abundant mucinous tumor tissue in the peritoneal cavity that eventually, if untreated, will compress and destroy the inner organs. PM is treated with cytoreductive surgery (CRS), followed by hyperthermic intraperitoneal chemotherapy (HIPEC) to kill the remaining tumor cells. The treatment is quite demanding with risk of complications, and the outcome is highly variable. Thus, finding biomarkers and new therapeutic targets is needed to facilitate treatment selection and discover new possible treatment options for these patients.
To approach this aim, we are performing multilevel molecular analysis on PM-CRC and PMP patient samples, harvested during surgery at the Norwegian Radium Hospital. DNA/RNA, extracted from blood and tumor samples, are subjected to next-generation sequencing, and through bioinformatics analysis (which is my main focus) we retrieve knowledge of the mutation profiles, copy number variations, gene expression and fusion genes common for these diseases. Associating these data with clinical characteristics will give us important knowledge for improving patient care (fig1).
Publications 2025
Enrichment of Cancer-Associated Fibroblasts, Macrophages, and Up-Regulated TNF-α Signaling in the Tumor Microenvironment of CMS4 Colorectal Peritoneal Metastasis
Cancer Med, 14 (1), e70521
DOI 10.1002/cam4.70521, PubMed 39739693
Publications 2024
Cytoreductive Surgery With Hyperthermic Intraperitoneal Chemotherapy and Liver Resection is a Treatment Option for Patients With Peritoneal and Liver Metastases From Colorectal Cancer
Ann Surg, 280 (5), 745-752
DOI 10.1097/SLA.0000000000006492, PubMed 39185557
Novel drug resistance mechanisms and drug targets in BRAF-mutated peritoneal metastasis from colorectal cancer
J Transl Med, 22 (1), 646
DOI 10.1186/s12967-024-05467-2, PubMed 38982444
Publications 2022
T cell receptor repertoire sequencing reveals chemotherapy-driven clonal expansion in colorectal liver metastases
Gigascience, 12
DOI 10.1093/gigascience/giad032, PubMed 37161965
A comprehensive framework for analysis of microRNA sequencing data in metastatic colorectal cancer
NAR Cancer, 4 (1), zcab051
DOI 10.1093/narcan/zcab051, PubMed 35047825
Publications 2021
Peptide vaccine targeting mutated GNAS: a potential novel treatment for pseudomyxoma peritonei
J Immunother Cancer, 9 (10)
DOI 10.1136/jitc-2021-003109, PubMed 34711663
Omics analyses in peritoneal metastasis-utility in the management of peritoneal metastases from colorectal cancer and pseudomyxoma peritonei: a narrative review
J Gastrointest Oncol, 12 (Suppl 1), S191-S203
DOI 10.21037/jgo-20-136, PubMed 33968437
The immuneML ecosystem for machine learning analysis of adaptive immune receptor repertoires
Nat Mach Intell, 3 (11), 936-944
DOI 10.1038/s42256-021-00413-z, PubMed 37396030
Publications 2020
Experimental Treatment of Mucinous Peritoneal Metastases Using Patient-Derived Xenograft Models
Transl Oncol, 13 (8), 100793
DOI 10.1016/j.tranon.2020.100793, PubMed 32447231
Publications 2019
Integrative genomic analysis of peritoneal malignant mesothelioma: understanding a case with extraordinary chemotherapy response
Cold Spring Harb Mol Case Stud, 5 (2)
DOI 10.1101/mcs.a003566, PubMed 30862609
Publications 2017
The rainfall plot: its motivation, characteristics and pitfalls
BMC Bioinformatics, 18 (1), 264
DOI 10.1186/s12859-017-1679-8, PubMed 28521741
GSuite HyperBrowser: integrative analysis of dataset collections across the genome and epigenome
Gigascience, 6 (7), 1-12
DOI 10.1093/gigascience/gix032, PubMed 28459977
Publications 2016
Hypoxia-induced alterations of G2 checkpoint regulators
Mol Oncol, 10 (5), 764-73
DOI 10.1016/j.molonc.2015.12.015, PubMed 26791779
Publications 2014
PLK1-inhibition can cause radiosensitization or radioresistance dependent on the treatment schedule
Radiother Oncol, 110 (2), 355-61
DOI 10.1016/j.radonc.2013.12.014, PubMed 24502970
Publications 2011
A genetic screen identifies BRCA2 and PALB2 as key regulators of G2 checkpoint maintenance
EMBO Rep, 12 (7), 705-12
DOI 10.1038/embor.2011.99, PubMed 21637299
The amount of DNA damage needed to activate the radiation-induced G2 checkpoint varies between single cells
Radiother Oncol, 101 (1), 24-7
DOI 10.1016/j.radonc.2011.05.060, PubMed 21722983
