Computational Systems Medicine in Cancer

Group leader
Introducing the group's aims (video)
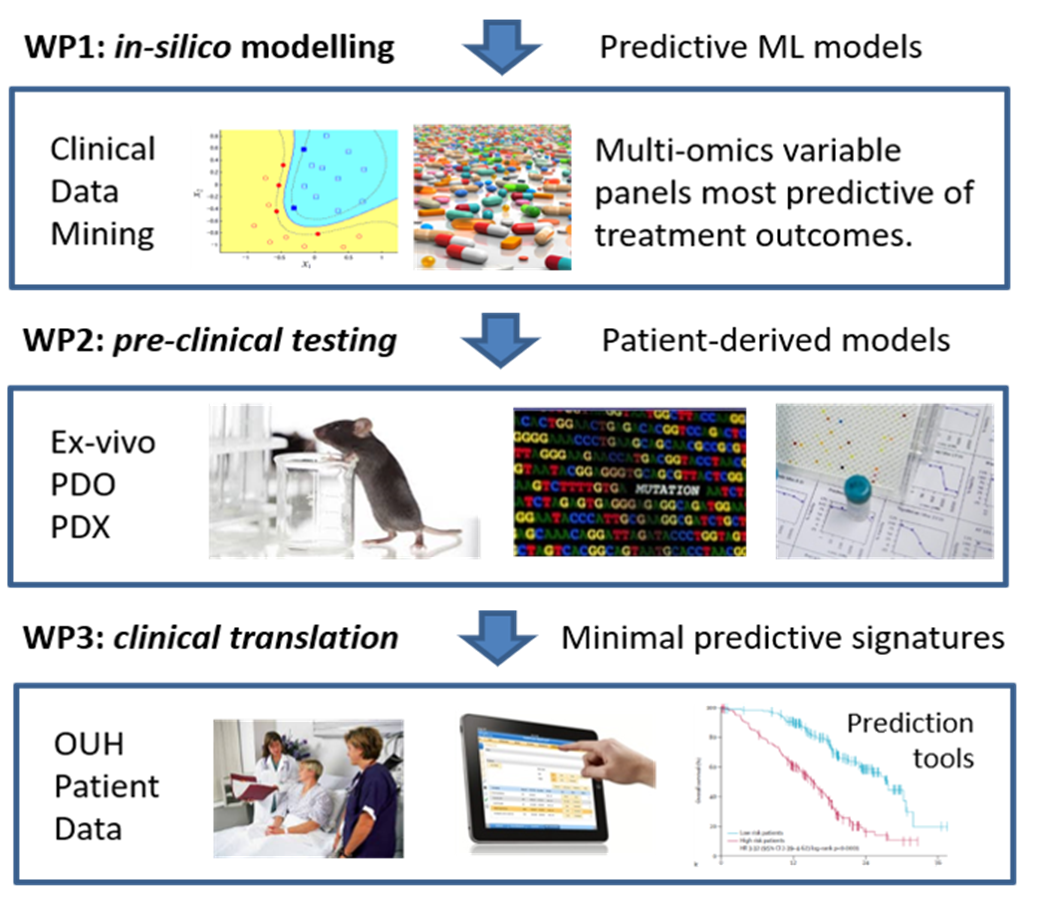
Our group has expertise in integrating multi-omics profiling and clinical information from cancer patients using mathematical and statistical approaches such as machine learning and network modeling. The medical aim is to optimize treatment outcomes for individual patients using maximally predictive models and minimal biomarker signatures that enable real-time and cost-effective routine diagnostics and prognosis. We believe that combining functional, molecular and genomic profiling information is critical for next-generation precision medicine applications, where integrative modeling and clever use of big data will pinpoint effective and selective targets for personalized therapies.