Blood plasma has been shown to carry small amounts of fragmented circulating cell-free DNA of tumour origin (ctDNA), which has been released to the blood stream through necrosis and apoptosis. These blood samples can be regarded as a non-invasive “liquid biopsies”, providing a real-time insight into the tumour’s genome, and can thus be used to monitor disease progression. By investigating ctDNA from cancer patients, the tumour heterogeneity can be better accounted for, since ctDNA in principle contains DNA representing every cancer cell within the body. As part of the Norwegian Cancer Genomics Consortium, our group collaborates with multiple groups in Norway to establish sequencing-based liquid biopsies analysis for research and clinical use.
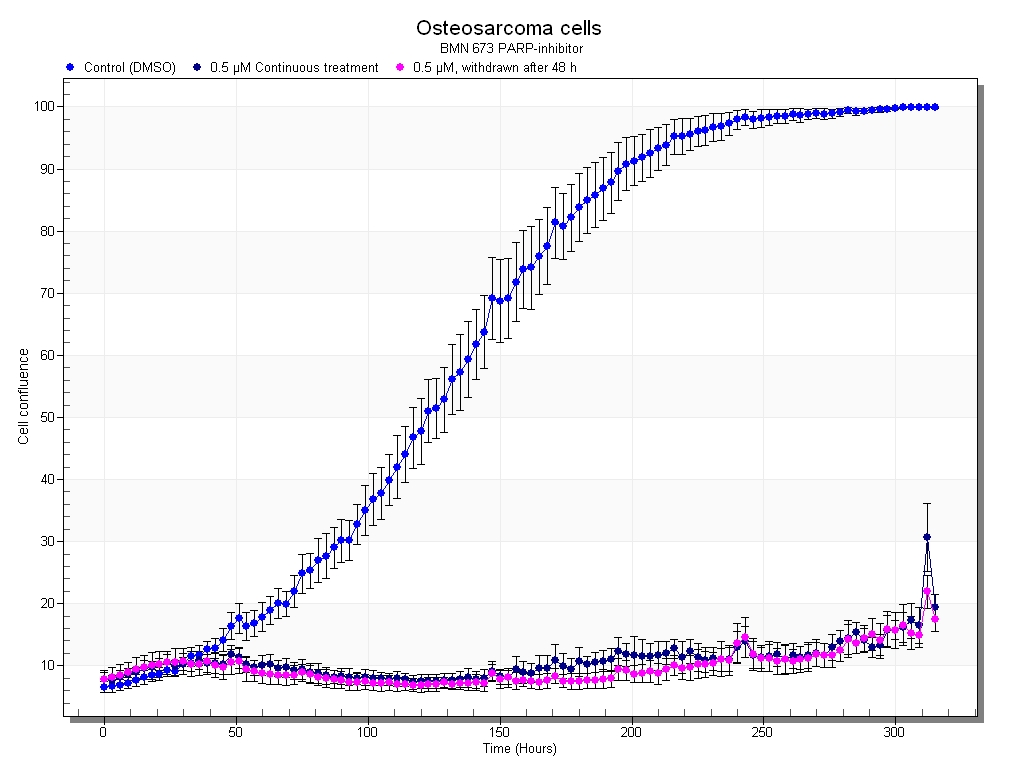
In 2014, our group has initiated a prospective study on circulating DNA in sarcomas (CircSarc). Plasma has been collected before and after surgery, and we are currently monitoring the patients over time, collecting plasma at routine controls and before and after each treatment cycle. This study aims to provide new insights into the clinical utility of liquid biopsies in soft tissue sarcomas, where somatic mutations in circulating cfDNA will be used to detect disease progression, monitor treatment response, study resistance mechanisms and possible therapeutic targets. Patients are begin recruited at Oslo and Haukeland University Hospitals.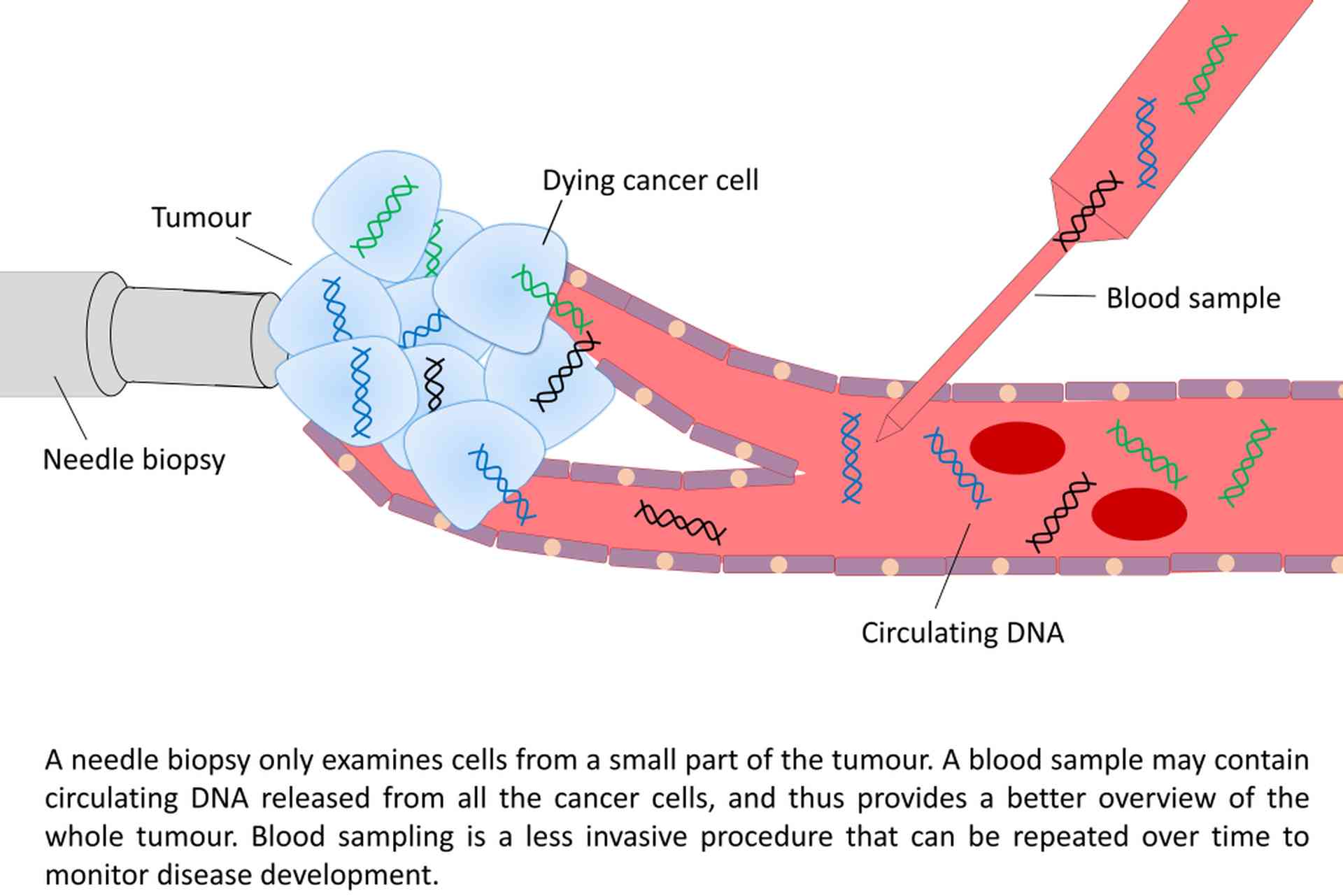
You can read more about the study in Sarkomer from the Norwegian sarcoma patient advocacy group.
A dedicated CircSarc project for gastrointestinal stromal tumours has been established with Dr. Boye at OUH in 2016, to analyse cfDNA from all recruited patients in the NoSarc study. In addition, longitudinal plasma samples are collected for metastatic patients and at selected time points during follow-up and before starting new treatments.
Our group is also establishing new sequencing-based technologies to detect RNA fusion transcripts in exosomes and platelets of cancer patients. Recent studies have shown that cancer cells can release microvesicles in the form of exosomes that might contain dsDNA and RNA. Similarly, “tumour-educated platelets” may contain tumour-derived RNAs. The aim of these studies is to show that cancer specific fusion transcripts can be successfully detected in the exosome or tumour-educated platelets in sarcomas, and further establish the use of fusion transcripts as a biomarker for non-invasive disease monitoring in fusion-driven sarcomas.