Leonardo A. Meza-Zepeda's project group Translational Genomics

The translational genomics group aims to understand cancer genome biology, by studying the genetic events that control cancer biology. Through advanced genomic analysis, we aim to identify and characterise novel biomarkers to better stratify sarcoma patients and ultimately translate this genomic knowledge to improve patient treatment and care.
Our group has pioneered the establishment of numerous high-throughput-based methods to study genome structure, dynamics and function in cancer, from bulk analysis to single-cell and spatial transcriptomics. We work loosely with the Genomics Core Facility, which I lead.
We work towards improving patient stratification of high-risk GIST in order to better control the disease and, through investigational studies, reveal novel therapeutic vulnerabilities in GIST undergoing imatinib treatment. This can be divided into the following projects.
Better risk stratification in GIST
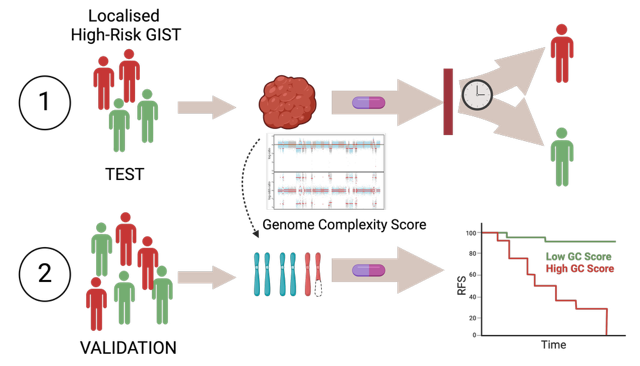
Risk stratification of GIST patients relies on clinical parameters such as tumour size, mitotic rate, tumour location, and tumour rupture to classify GISTs into very low–, low-, intermediate-, and high-risk. Today, adjuvant treatment with imatinib for three years is recommended for patients with localised GIST who have a significant risk of relapse. Patients classified as high-risk benefit from adjuvant imatinib treatment, but around 40% of patients relapse within three years after stopping adjuvant treatment. Furthermore, historical data has shown that almost half of the high-risk patients could be cured by surgery alone. Thus, the high-risk group includes both tumours with a good and bad prognosis that may or may not benefit from imatinib treatment. Thus, there is a clinical need to better stratify high-risk GIST patients to identify, at a diagnostic time point, which patients may benefit from a more prolonged imatinib treatment, and which will not.
Using a multi-omics approach of untreated high-risk GIST which progressed rapidly after adjuvant imatinib treatment or showed prolonged stable disease, we have identified DNA copy number aberrations (CNA) and loss of heterozygosity (LOH), seen as high genome complexity, as an intrinsic feature in tumours with more aggressive behaviour (Namløs et al, Mol Oncol, 2023). We are further validating the use of genome complexity as a novel prognostic biomarker for high-risk GIST.
Tumour Heterogeneity and Drug Resistance
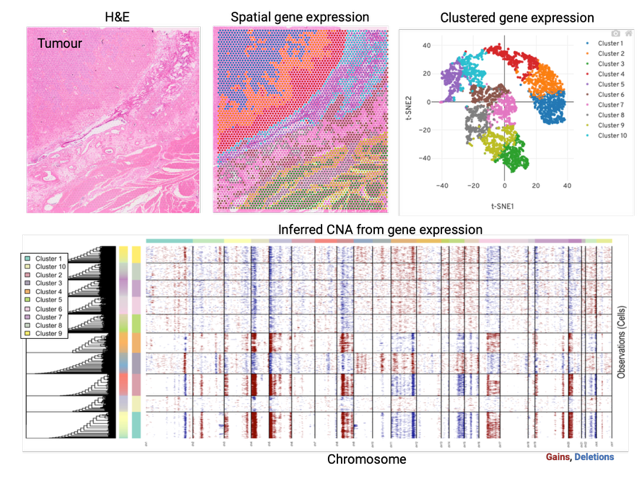
Disease progression is a major issue for successful cancer management. Understanding the molecular determinants of progression is essential for the development of effective and durable therapeutic strategies. The ability of cancer cells to adapt to treatment can be fuelled by the intrinsic heterogeneity of cancer cells within a tumour. We work to characterise the genomic and transcriptional heterogeneity in treatment naïve GIST and correlate this to response to adjuvant treatment. This is performed by spatial transcriptomics analysis using a selected cohort of GIST clinical samples.
Liquid Biopsies
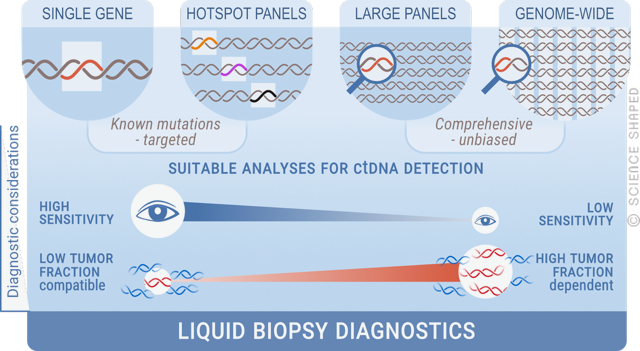
The blood plasma of cancer patients carries small amounts of fragmented circulating cell-free DNA (cfDNA) of tumour origin (ctDNA), which are released into the bloodstream. The ctDNA represents, in principle, every cancer cell within the body, and using a blood sample as a “liquid biopsy” allows non-invasive means to obtain tumour material for molecular analyses, thus circumventing the limitation of traditional biopsies. Our group has established a number of high-throughput sequencing methods to study liquid biopsies, which are used to monitor treatment response, disease evolution, and drug resistance in different types of soft tissue sarcomas, including GIST. We aim to develop a less invasive and more sensitive way of real-time monitoring of disease.
We also participate in different collaborative efforts to implement advanced genomic testing of cancer patients to drive precision cancer medicine in Norway. One of these efforts is InPreD, an infrastructure for precision diagnostics, which delivers advanced diagnostics to stratify cancer patients into clinical trials.
Our research is financed by the Norwegian Cancer Society

I'm also head of the Helse Sør-Øst and University of Oslo Genomics Core Facility and head of the NorSeq-Cancer node of the National Technology platform for Sequencing and Personalised Medicine. I also lead the Department of Core Facilities at the Institute for Cancer Research, OUH.